
Save time at the bench - no bacterial gDNA extraction is required. Simply submit your clonal bacterial sample, and GENEWIZ will perform PCR amplification and Sanger DNA sequencing using 16S primers. GENEWIZ sequences the 16S rRNA gene from your bacterial colonies rapidly and effectively. Due to this conservation, scientists can use the 16S rRNA gene to identify prokaryotic species present in a sample. Due to the structural importance of this gene, the pace of evolution of the gene has been relatively slow. The 16S ribosomal RNA subunit is an essential component in the 30S ribosomal complex in prokaryotes. 16S rDNA sequencing analysis is characterized by high sequencing flux, large amount of data obtained, short cycle, and more comprehensive reflection of. Azenta Life Sciences Consumables & Instrumentsġ6S rRNA: Let GENEWIZ Help Construct Your Phylogenies.Gene Synthesis & Cloning/Mutagenesis FAQs.CLIA Variant Confirmation (PCR + Sanger).The 16S workflow will be useful to any researcher interested in identifying pathogens in a mixed sample or understanding the composition of a microbial community both already thriving subjects of research within the Nanopore Community.
16s rdna sequence analysis full#
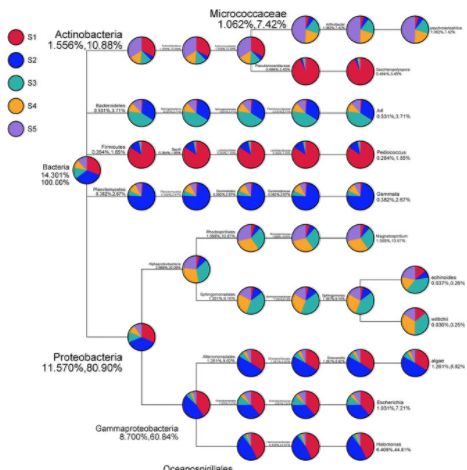
Each read is classified based on % coverage and identity. 16S and Internal Transcribed Spacer (ITS) ribosomal RNA (rRNA) sequencing are common amplicon sequencing methods used to identify and compare bacteria or fungi. The MicroSEQ Full Gene 16S rDNA Sequencing Kit is the sequencing component of the MicroSEQ 16S rDNA Full Gene Identification System, which provides an. The workflow is designed to BLAST basecalled sequence against the NCBI 16S bacterial database, which contains over 20,000 16S sequences from different organisms. Oxford Nanopore also provides a complete 16S analysis workflow, enabling users to classify mixed samples and obtain accurate results for single reads at the genus level. Between the WIMP and 16S workflows, nearly all scenarios for taxonomic assignment of small-genome species are covered. This is in contrast to the What's In My Pot (WIMP) application, which aligns all reads from a mixed metagenomic sample to an NCBI database of bacteria, viruses, fungi, and archaea. Metagenomics application page, containing white papers, videos, and peer-reviewed publications A total of 43 bacterial strains were selected based on colony. In this situation, where a large number of species occur and many species are unculturable, phylogenetic analysis of sequence data from the 16S ribosomal. Further resourcesġ6S Workflow: see tutorial video in the Nanopore Community (login required) The sequence analysis of the small subunit, 16S rDNA, has been frequently used for determining inter- and intraspecific relationships. The objective of this study ws to use 16S rDNA sequence analysis to better identify WDS bacteria. fastidiosa strains showed a maximum variation of 1.0 or 14 nucleotide positions. Furthermore, analysis of the full-length 16S rDNA gene using long-read nanopore sequencing has been shown to enhance the resolution of microbial characterisation. Abstract The 16S rDNA encoding the small subunit ribosomal RNA were amplified by PCR, cloned, and sequenced from 16 strains of Xylella fastidiosa originating from nine different hosts. information, the 16S rDNA sequences containing internal. 16S rRNA gene sequence analysis can better identify poorly described, rarely isolated, or phenotypically aberrant strains, can be routinely used for. By narrowing down to a specific region of interest, all the bacterial genera in the sample can be seen without sequencing unnecessary regions of the genome and avoiding problems of host contamination, making the test quicker and more economical. reported a classification analysis based on the 16S rRNA sequence of an organism formerly. The MinION has been used by many researchers for 16S analysis.
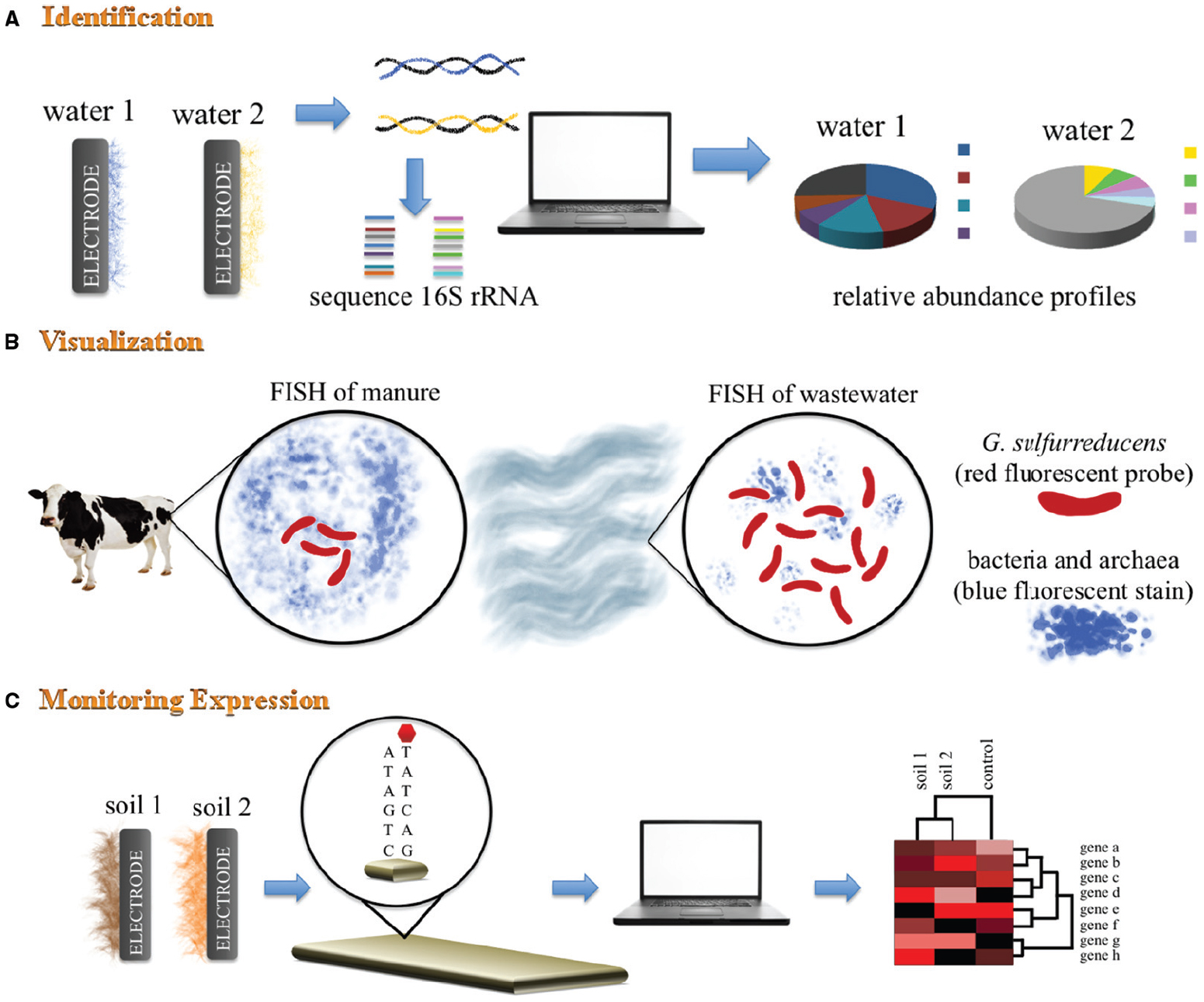
Notably, the gene has two different domains, a larger one conserved domain, and a hypervariable region.
Although the gene is highly conserved since evolution and therefore it is often known as a molecular fossil. In addition, the 16S Barcoding Kit and 16S Barcoding Kit 1-24 provides 12 and 24 barcodes respectively, for multiplexing up to 12 or 24 samples, enabling even more cost-effective results. 16S rRNA gene constructs the 16S rRNA subunit which binds to the Shine-Dalgarno sequence present in the bacteria genome. Note: Sample preparation components not included in the kit. 16s rrna sequencing is an affordable method. The 16S Barcoding Kits and EPI2ME 16S analysis workflow allows users to perform genus-level identification from single reads with access to basecalled files for detailed investigations at the species and sub-species level. The MRDNA in-house 16srrna sequencing primers are able to target the hypervariable regions of the 16s rrna gene.


 0 kommentar(er)
0 kommentar(er)
